AI to Interpret Your Mind
What if a machine learning model could interpret what’s going on inside our minds?
We could all use more reasons to be optimistic, so I’ve started a project called precisely that.
Going forward, I’ll be sharing deep dives on the technologies I believe everyone should pay attention to, but perhaps aren’t. Think low-level enough to interest engineers, but high-level enough for everyone to grasp why it matters.
There has been some remarkable progress in our ability to reconstruct the thoughts going on inside our own head. The technology today has significant limitations and is far from mature, but it is a glimpse into a future where our mind alone could trigger actions outside our bodies. If we fast forward the clock a decade or two, what might be possible?

Above we can see two columns of images. The image on the left—the “seen image”—is a picture that is physically shown to a human. Then, the image on the right—the “reconstruction”—is what a ML model guesses the human saw using just a brain scan as the input. The first reaction of many to this is disbelief. How is it possible that a ML model can interpret what we saw from just a brain scan, and reconstruct such eerily accurate images?
In this post, we’ll go deeper on how these results were achieved and the state-of-the-art in brain image reconstruction. But, first let’s understand the brain, the tools to image it, and the history of those trying to better understand it.
The Human Brain
The brain controls all complex functions in the body, and is the center where our thoughts and experiences are processed. Since the brain is organized into structural groups, if we can track the activations of neurons, we can attempt to map what’s going on inside the brain. Thoughts, movements, and your biological control processes could be analyzed and recovered. This is the holy grail that many in cognitive neuroscience have been working toward.
To interface with the brain, there are two main approaches. Invasive measures involve directly interacting with the brain tissue within the skull, with things like probes and wires. Noninvasive measures involve using electromagnetic fields and radiation to map the brain, without actually touching the brain. For this newsletter, we are diving into the development of noninvasive technology.
There’s been some mind-blowing developments in this space. So, to appreciate the long journey that brought us here, let’s dive into the history of noninvasive brain imaging.
The History of Noninvasive Brain Scanning
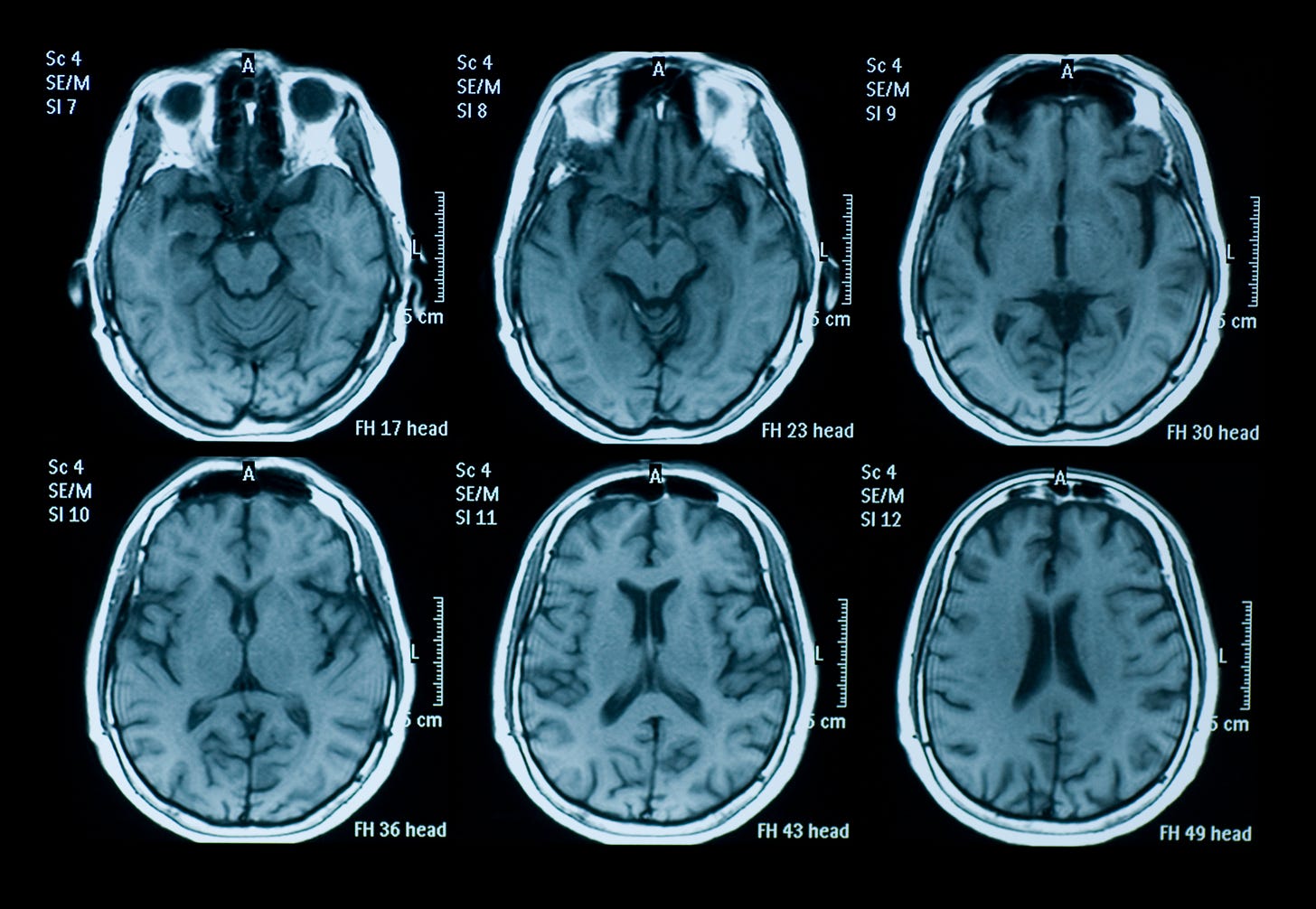
The first hardware breakthrough to noninvasive brain imaging was the invention of the Magnetic Resonance Imaging (MRI) machine in the early 1970s. The foundational principles behind MRI were first described by physicists Paul Lauterbur and Sir Peter Mansfield, who independently developed techniques to produce detailed images using magnetic resonance. Their work earned them the Nobel Prize in Physiology or Medicine in 2003.
MRIs export a 3D scan of the structure of the brain. They work by using powerful magnets that align all protons in the body to the field. Radio current is pulsed through the patient, which moves protons in the brain out of equilibrium. When the field is turned off, MRI sensors detect the energy released, and the time it takes for the protons to realign. Different types of tissues have different molecular properties, so we can build a 3D map of structural components of the brain based on this information.
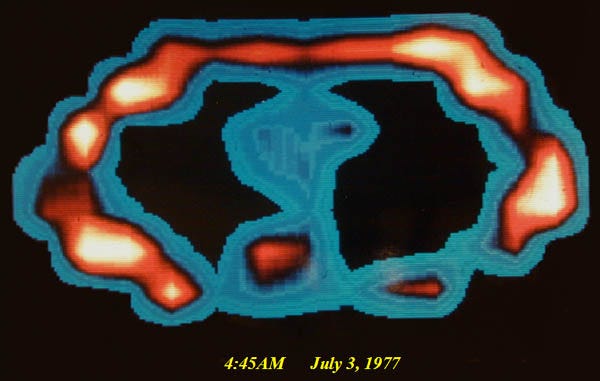
MRIs have been a huge boon to the medical field, providing an indispensable tool for the diagnosis of all brain-related diseases. In the 1990s, Seiji Ogawa and Ken Kwong helped develop the Functional MRI (fMRI). This builds upon MRIs, but enables measuring the activity of the brain in real-time, rather than just the physical structure of the tissues within the brain.
fMRI tracks neural activity by measuring blood flow. It’s based on a physiological phenomenon called the hemodynamic response. Neurons do not contain internal reserves of glucose or oxygen, so blood flow is directed on demand to active regions of the brain. fMRIs use blood oxygen level-dependent (BOLD) contrast imaging to measure the levels of oxyhemoglobin and deoxyhemoglobin based on their magnetic properties, which give us a near real-time picture of which neurons are firing in the brain.
Almost all noninvasive brain reading projects use fMRI technology, due to it being far more precise than alternatives such as EEG or MEG. However, one of the many downsides of fMRIs is that they are extremely expensive, and to put it lightly, large.
Ultimately, these downsides have resulted in very limited sets of data to work with. In addition, although fMRI is currently the most precise source, it still has a low signal-to-noise ratio, due to variables such as patient movement, and the fact that every brain has slightly different structures and blood flow patterns.
The History of Brain Image Reconstruction
Since the advent of the fMRI, there has been an explosion of research work in interpreting brain activity from fMRI scans. This is a quickly growing field, with more than 1,000 research papers and counting. We’ll dive into image reconstruction specifically: the task of reconstructing what the patient saw in real-life from brain scan data.
In each of these experiments, a patient is shown an image inside an fMRI machine. The goal is to then reconstruct that shown image from the fMRI scan itself. An fMRI scan consists of a 3D matrix of pixels, which are called voxels. Each voxel has a discrete value, which represents how much blood is in the 3D volume occupied by the voxel.
Since the early 2000s, there has been a constant stream of research work involving using statistical methods to make sense of the pattern and structure of fMRI scans in the visual cortex. Bayesian statistics is a common technique in many approaches, which involves calculating the probability of some neural activity (i.e. the visual cortex seeing a cat) based on the given event (an fMRI scan).
One of the first landmark achievements was a paper by Kamitami et al in 2005. The team hypothesized that each voxel could contain some weak information about the stimulus, and that if processed properly, the sum total information of all the voxels in a scan could reveal some information about the stimulus the subject was experiencing. So, by using simple linear regression on each individual voxel, Kamitani et al were able to predict the amount of rotation of an image that was shown to the subject. They were quite good at achieving this, with a 2-way accuracy level of around 80%.
Kamitami et al’s work is significant because it’s a trailblazer in using machine learning to recover details about a person’s visual experience. It proved that we can interpret part of the mind to a degree, even if it’s just to see how much an image is rotated.
The next breakthrough was Kay et al’s 2008 paper, which used linear regression to attempt to predict what image a person was shown. This is an example of a “pick a card, any card” approach, where someone shows you an image from a predefined set of images, and tries to figure out what image you were shown. This includes novel images, meaning cards that they haven’t seen before. Understandably, you might think this is a bit crazy—how can you predict images that you didn’t train on? The trick is to train a receptive-field model that describes how image properties (such as patterns, shapes) affect each fMRI voxel.
This approach was achieved like so: Kay et al first showed people images, and recorded their fMRI for those images. Using this dataset, they trained their receptive-field model. For each image in the test dataset, they used the receptive-field model to predict the fMRI activations of each image. At test time, they show a person an image and measure their BOLD response using the fMRI. Then, they see which predicted fMRI activation matches the most with the ground truth fMRI, and predict the image that corresponds to that. They achieved 92% accuracy on one subject and 72% on another subject with P < 0.008. Note that there’s some variability between subjects, which is natural due to slight differences in their brain structure, hemodynamic response, and other experimental variables like their position in the fMRI machine. It’s also noteworthy that Nishimoto et al. used the same approach for videos in 2011.
Come 2018, we are seeing some exciting new tools in machine learning. Convolutional Neural Networks (CNNs) were performing well, because their architecture was inherently optimized to work with image data. The idea was; what if we learn the “meaning” of images rather than just their statistical attributes? Thus, the idea of CNNs is to capture both low and high levels features of an image by convolving the pixels using kernels. This maintains more semantic and structural features of the image rather than predicting each pixel separately, as was traditionally done with linear regression.
Wen et al. used deep CNNs directly on fMRI signals to accomplish two tasks: predict object classes and directly recreate the images. First, they trained a CNN to predict classes from movie frames. Then, they trained a linear regression model to take a late CNN layer (which contains higher level semantic details) to predict fMRI activity. At test time, they would run an fMRI backwards through the model.
When predicting the category of the image from one of 15 categories (i.e. people, bird, airplane), the top 1 accuracy score was an average of 48%. This was a breakthrough, as it was one of the first projects to not only predict which type of image you saw, but also tried to reconstruct what it looked like. Its use of a late-stage CNN layer was also a baby step towards the idea of an fMRI latent (lower dimensional vector) to predict images.

So far, projects relied on a very small set of image and fMRI pairs, which made it hard to train powerful models. There were not enough labeled examples, simply because fMRIs scans are notoriously expensive to perform at scale.
Come Beliy et. al to the rescue in 2019, who introduced the idea of unsupervised learning to image reconstruction. They used supervised data to train an image to fMRI encoder, and an fMRI to image decoder. Essentially, they were regular CNNs that mapped from one domain to another. They then connected the encoder to the decoder back-to-back, allowing self-supervised training on 50,000 ImageNet images. Then, they connected the decoder to encoder back-to-back, allowing self-supervised training on unlabelled test fMRI recordings, as shown in the diagram below.
This suddenly drastically increased the amount of data that was available to train the model. Finally, taking just the decoder, they were able to yield some impressive results…
Recent Progress in Brain Image Reconstruction
The past few years have been a time of remarkable progress in the field. Thanks to the introduction of transformers, and generative models such as diffusion models, we’ve seen some leapfrog improvements in reconstruction quality.
The two main avenues of improvement were through learning fMRI representations better and superior image reconstruction. With the advent of transformers, researchers began using autoencoders to learn the meaning of labeled and unlabeled fMRI scans. In 2022, we also saw a few exciting papers that used Stable Diffusion to generate images.
In 2022, Lin et al. mapped fMRI latents to the CLIP space, which increased accuracy by 3-4%. They also generated images from these latents with a GAN, and were the first to reconstruct complex images.
In late 2022, Chen et al. created Mind-Vis, which used a masked vision autoencoder to build a better fMRI latent learner. They also used a diffusion model instead of a GAN for better generation quality.
The architecture for this new generation of image reconstruction projects usually consists of two parts: an fMRI learner and an image generator.
So far, diffusion-based projects generated images from pure noise. What if, instead of diffusing from noise, we could diffuse from a fuzzy image that matches the low-level structural shapes of the image? This would combine the pixel-level performance of Beliy et al’s work with the semantic-level performance of Chen and Lin et al’s work.
Scotti et al did this through MindEye, which achieved some breathtaking results.
Brain Video Reconstruction
Thanks to increased attention from the public and academia, there is a recent surge of growth in the fMRI reconstruction space. The new projects aim to improve on performance, interpretability, and multi-modality.
Let’s dive into a new project, Mind-Video, by Chen et al. This is the first recent project that works with video: subjects are shown a video, and the model attempts to reconstruct that video.
It builds upon the architecture of Mind-Vis, but with a few modifications. First, it uses classifier-free guidance, which essentially passes stronger embeddings to the Stable Diffusion model for a more specific reconstruction. They also changed the architecture to work with video frames instead of images. This is quite a technical challenge, as fMRI scans capture BOLD changes over time, which gets jumbled over time.
Diving deeper into the architecture, it involves first pretraining a ViT-based masked auto-encoder on the Human Connectome Project dataset, which is a great source of unlabelled fMRI data. Then, the SC-MBM encoder is fine tuned on an fMRI-video dataset introduced by H. Wen et al in 2018. We want to project the learned fMRI latent to the same space as the latent of CLIP based image encoder and CLIP based text encoder, as the diffusion model used in the generative step takes in cross attention latents in the CLIP space.
So, the team trained the SC-MBM encoder to map the latents to the image and text latents by minimizing the cross entropy loss between the three latents, where only the fMRI encoder weights were not frozen. The trick here is that one fMRI actually corresponds to 6 frames of video data, and is actually delayed in time due to the hemodynamic response. To address this, the team paired fMRIs with video frames shifted a bit forward in time.
The team then passed the latents to a Stable Diffusion architecture adapted to video, introduced in “One-shot tuning of image diffusion models for text-to-video generation” by J. Z. Wu et al. It works by essentially stacking frames of a video on top of each other, to create a 3-dimensional matrix.
All in all, this is a very exciting result, because we are seeing the early potential to capture “multi-frame” thoughts and experiences from brain scans. The implications of this is that it may be feasible to transcribe continuous data from brains, and possibly in real-time as well.
What Might Be Possible?
Brain image reconstruction potentially enables whole new paradigms of understanding and communication. However, we should be wary of assigning too much weight to the accuracy of reconstructed images, as fMRI data is noisy, and current methods such as Stable Diffusion rely on are probabilistic in nature. Basically, we are very early on this exciting journey.
But, if we dream, what are some possibilities?
In the future, what if you could control the physical world or send messages just by thinking, all without a brain implant? Or, what if your thoughts could be privately recorded for you to remember and search through later? What if you could analyze and share dreams? What if we could investigate differences in perception across people, to map phenomena such as personality and attention?
This reality sounds impossible today, especially noninvasively, and it may ultimately prove to be. But, thanks to decades of work in this field, and the trajectory of current research, I believe there are reasons to be optimistic!
What Happens Next?
Reach out to Jonathan Xu if you’re a researcher, engineer, or investor, and would like to collaborate. He’s currently working on the second version of Mind-Vis, implementing some innovations to greatly improve its capabilities. He’s also compiled some resources if you'd like to explore fMRI datasets and opportunities. We’re excited for where this is leading, including learning rich fMRI latents incorporating spatial information, and adapting the architecture to more approachable hardware such as MEGs and EEGs. Thanks for reading!
Sources
Kamitani, Y. & Tong, F. Decoding the visual and subjective contents of the human brain. Nature Neurosci. 8, 679–685 (2005)
K. N. Kay, T. Naselaris, R. J. Prenger, and J. L. Gallant, “Identifying natural images from human brain activity,” Nature, vol. 452, pp. 352–355, 3 2008.
H. Wen, J. Shi, Y. Zhang, K.-H. Lu, J. Cao, and Z. Liu, “Neural Encoding and Decoding with Deep Learning for Dynamic Natural Vision,” Cerebral Cortex, vol. 28, pp. 4136–4160, 12 2018.
Roman Beliy, Guy Gaziv, Assaf Hoogi, Francesca Strappini, Tal Golan, and Michal Irani. From voxels to pixels and back: Self-supervision in natural-image reconstruction from fMRI. Advances in Neural Information Processing Systems, 32, 2019.
Lin, Sikun, Thomas Sprague, and Ambuj K. Singh. "Mind Reader: Reconstructing complex images from brain activities." arXiv preprint arXiv:2210.01769 (2022).
Chen, Zijiao, et al. "Seeing beyond the brain: Conditional diffusion model with sparse masked modeling for vision decoding." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2023.
Lv, Han, et al. "Resting-state functional MRI: everything that nonexperts have always wanted to know." American Journal of Neuroradiology 39.8 (2018): 1390-1399.
Chen, Zijiao, Jiaxin Qing, and Juan Helen Zhou. "Cinematic Mindscapes: High-quality Video Reconstruction from Brain Activity." arXiv preprint arXiv:2305.11675 (2023).
Scotti, Paul S., et al. "Reconstructing the Mind's Eye: fMRI-to-Image with Contrastive Learning and Diffusion Priors." arXiv preprint arXiv:2305.18274 (2023).